Healthcare-associated infection (HAI) - March 30, 2023
The different C. difficile diagnostic algorithms and the impacts on molecular diagnostics
Clostridioides difficile: prevalence and pathophysiology
C. difficile is a spore-producing, gram-positive bacterium most associated with nosocomial antimicrobial-associated diarrhea.1 The anaerobic microbe is found in the lower digestive
 system of approximately 20-40% of hospitalised adults compared with 2-3% or 1 in 30 healthy adults.2,3 C. difficile does not cause disease in healthy individuals.3,4 The disease is facilitated by the production of enterotoxin (A) and cytotoxin (B), and infection only occurs when C. difficile becomes established in the gastrointestinal (GI) tract when the normal population of enteric bacteria is disrupted, usually due to antibiotic exposure.1,3
system of approximately 20-40% of hospitalised adults compared with 2-3% or 1 in 30 healthy adults.2,3 C. difficile does not cause disease in healthy individuals.3,4 The disease is facilitated by the production of enterotoxin (A) and cytotoxin (B), and infection only occurs when C. difficile becomes established in the gastrointestinal (GI) tract when the normal population of enteric bacteria is disrupted, usually due to antibiotic exposure.1,3
However, C. difficile infections (CDIs), all be it preventable, occur regularly and pose a significant risk to patient mortality and morbidity, particularly in those with antibiotic exposure, advanced age, prior hospitalization, those who use feeding tubes or have underwent gastrointestinal surgery.5 In Europe, CDIs account for approximately 60% of gastrointestinal infections, which comprises approximately 9% of all healthcare-associated infections (HAIs).6 The European Society of Clinical Microbiology and Infectious Diseases (ESCMID) identified one in ten cases of CDI cause, or contribute to, intensive care unit (ICU) admission or lead to a colectomy or death.6
C.difficile is easily spread in a hospital environment as the bacterium spores in a patient’s faeces can survive for an indefinite period.3 The spores are easily transferred to other patients via the faecal-oral route, via the hands of healthcare workers and contaminated surfaces.3 Additionally, the commonly used alcoholic antibacterial hand gels are ineffective against the persistent spores, requiring soap and water to eliminate them from a surface.3
Patients present with symptoms ranging from mild to severe diarrhea to life-threatening bowel complication, including toxic megacolon, pseudomembranous colitis, and septic shock.6 In addition to the impact on patients, CDIs account for a substantial burden to healthcare resources and costs partly due to extended periods of hospitalisation of up to 3 weeks.6
Where does the problem lie?
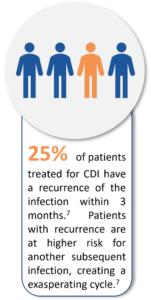
Firstly, CDIs have increased in prevalence and there has been significant changes in their clinical presentation, including more severe disease.7 In Europe, the estimated number of cases is 12,4000 per year with C. difficile reported to be the sixth most frequent microorganism responsible for HAIs between 2016 and 2017.8
Secondly, laboratory testing alone cannot distinguish between asymptomatic colonisation and symptomatic CDI.9 Furthermore, transmission risk is increased as many patients admitted to hospital are asymptomatic carriers of C. difficile acting as a potential infection reservoir.10
Multiple diagnostic tests can be used in various combinations for the diagnosis of CDIs.9 They can be used to detect C. difficile-specific nucleic acid, enzyme, and/or toxins. As a result, there are multiple diagnostic algorithms available, thus leading to considerable confusion regarding clinical interpretation and which algorithm detects the most cases whilst remaining cost effective.
Why are multiple tests needed for optimal diagnosis?
Testing is challenging from an infection prevention perspective; if the sensitivity of the testing strategy is too low, there is a higher risk of transmission. If the testing strategy has low specificity, a substantial proportion of patients are placed in isolation unnecessarily. It is generally accepted that positive assays for C. difficile toxins are indicative of active disease and that the toxins alone are responsible for the clinical symptoms.10 However, it has been reported that patients with positive toxin assays can remain symptomless.10 Therefore, the sole presence of toxins is insufficient for a diagnosis of the disease. A positive diagnosis requires the presence of three or more unformed stools in 24 or fewer consecutive hours and either: a positive stool test for toxins, a detection of toxigenic C. difficile, or colonoscopy findings showing pseudomembranous colitis.10
Historically, the gold standard for testing has been toxigenic culture from stool. Isolates are then tested for their ability to produce toxin A and B.11 Additionally, stool filtrates can be directly tested for the toxin via a cell cytotoxicity assay which acts as an alternative reference method.11 Unfortunately, these methods are time consuming, taking several days to complete, which is less than suitable for a high testing turnaround requisite in a hospital setting. Toxin detection by qualitive enzyme immunoassays (EIAs) used to be a mainstay of diagnosis; however, these assays have significant limitations in sensitivity compared to toxigenic cultures (52-75%).11 Nevertheless, EIAs have higher specificity (96-98%) when comparing to cultures.11
Glutamate dehydrogenase (GDH) immunoassays detect the metabolic enzyme present in all C. difficile isolates, yet this antigen is present in both toxigenic and nontoxigenic strains.12 This means that a more specific molecular test is needed for toxin gene detection. Notably, the discovery of C. difficile isolates that produce toxin B but not toxin A have indicated that the presence of toxin A is not essential for CDI onset.13 Strains producing only toxin B are equally as virulent as strains producing both toxins in human patients.14 Nucleic acid application tests (NAATs) are recommended by ESCMID guidelines as a primary test in a 2-step algorithm.12 These assays are more sensitive than toxin EIAs and GDH EIAs, but less sensitive than toxigenic cultures.12
No single commercial test can be used as a stand-alone test for diagnosing CDI as a result of positive predictive values at low CDI prevalence.12 Therefore, the two-step algorithm has been recommended by ESCMID to decrease the possibility of false positive results.12,15 The two-step process begins with the use of a high sensitivity test, either NAAT for detection of the toxin gene or an enzyme EIA to detect GDH, followed by highly specific tests that identifies free faecal C. difficile toxin, i.e., toxin EIA. In cases of negative results from the toxin test, NAAT (if not already the primary test) or toxigenic culture can be performed based on clinical evaluation. It is important to remember that a diagnosis of CDI is based on a combination of laboratory results plus positive clinical signs.
Testing in practice
A European study conducted by Le Guern and colleagues assessed the suitability of two molecular assays.16 The two assays were evaluated against an EIA followed by a toxigenic culture (TC).16 The results showed that the automated extraction and real-time PCR amplification process leaves the hands-on time necessary to process 10 samples at just 10 minutes.16 In terms of sensitivity, the EIA presented a low sensitivity at 43.2% whereas the 2 NAATs presented at 97.7% and 95.5% .16 In comparison, a 2018 study concluded that the sensitivity of a GDH and toxin EIA based algorithm was 85%, implying that cases would be missed 15% of the time.17 While the sensitivity of NAATs was 88-94%.14
References
- Mada PK, Alam MU. Clostridium Difficile.; 2021. Available at: http://www.ncbi.nlm.nih.gov/pubmed/28613708. Accessed November 2021
- Heinlen L, Ballard JD. Clostridium difficile Infection. The American Journal of the Medical Sciences 2010;340:247-252. Available at: https://linkinghub.elsevier.com/retrieve/pii/S0002962915315056.
- NHS. Clostridium difficile. NHS 2018. Available at: https://www.nhs.uk/conditions/c-difficile/. Accessed November 2021
- NHS. MRSA. NHS 2020. Available at: https://www.nhs.uk/conditions/mrsa/. Accessed November 2021
- Lin HJ, Hung YP, Liu HC, et al. Risk factors for Clostridium difficile-associated diarrhea among hospitalized adults with fecal toxigenic C. difficile colonization. Journal of Microbiology, Immunology and Infection 2015;48:183-189.
- Barbut F, Cornely O, Fitzpatrick F, et al. Clostridium difficile infection in Europe.; 2013. Available at: http://www.multivu.com/assets/60637/documents/60637-CDI-HCP-Report-original.pdf. Accessed November 2021
- Wiuff C, Banks A-L, Fitzpatrick F, et al. The Need for European Surveillance of CDI. In: Advances in Experimental Medicine and Biology.Vol 1050. Springer New York LLC; 2018:13-25. Available at: http://link.springer.com/10.1007/978-3-319-72799-8_2.
- Guery B, Galperine T, Barbut F. Clostridioides difficile : diagnosis and treatments. BMJ 2019;366:l4609. Available at: https://www.bmj.com/lookup/doi/10.1136/bmj.l4609.
- Bagdasarian N, Rao K, Malani PN. Diagnosis and Treatment of Clostridium difficile in Adults. JAMA 2015;313:398.
- Furuya-Kanamori L, Marquess J, Yakob L, et al. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infectious Diseases 2015;15:516.
- Lee R. C. diff Diagnosis versus Detection: Why Tests Remain Ambiguous. American Society for Microbiology 2019. Available at: https://asm.org/Articles/2019/January/C-diff-Diagnosis-versus-Detection-Why-Tests-Remain. Accessed November 2021
- Crobach MJT, Planche T, Eckert C, et al. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clinical Microbiology and Infection 2016;22:S63-S81.
- Drudy D, Fanning S, Kyne L. Toxin A-negative, toxin B-positive Clostridium difficile. International Journal of Infectious Diseases 2007;11:5-10.
- Awad MM, Johanesen PA, Carter GP, et al. Clostridium difficile virulence factors: Insights into an anaerobic spore-forming pathogen. Gut Microbes 2015;5:579-593.
- Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. The Lancet Infectious Diseases 2013;13:936-945.
- Le Guern R, Herwegh S, Grandbastien B, et al. Evaluation of a new molecular test, the BD Max Cdiff, for detection of toxigenic clostridium difficile in fecal samples. Journal of Clinical Microbiology 2012;50:3089-3090.
- Gomez EJ, Montgomery S, Alby K, et al. Poor yield of Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays in a pediatric population with declining prevalence of clostridium difficile strain BI/NAP1/027. Diagnostic Microbiology and Infectious Disease 2018;91:229-232.